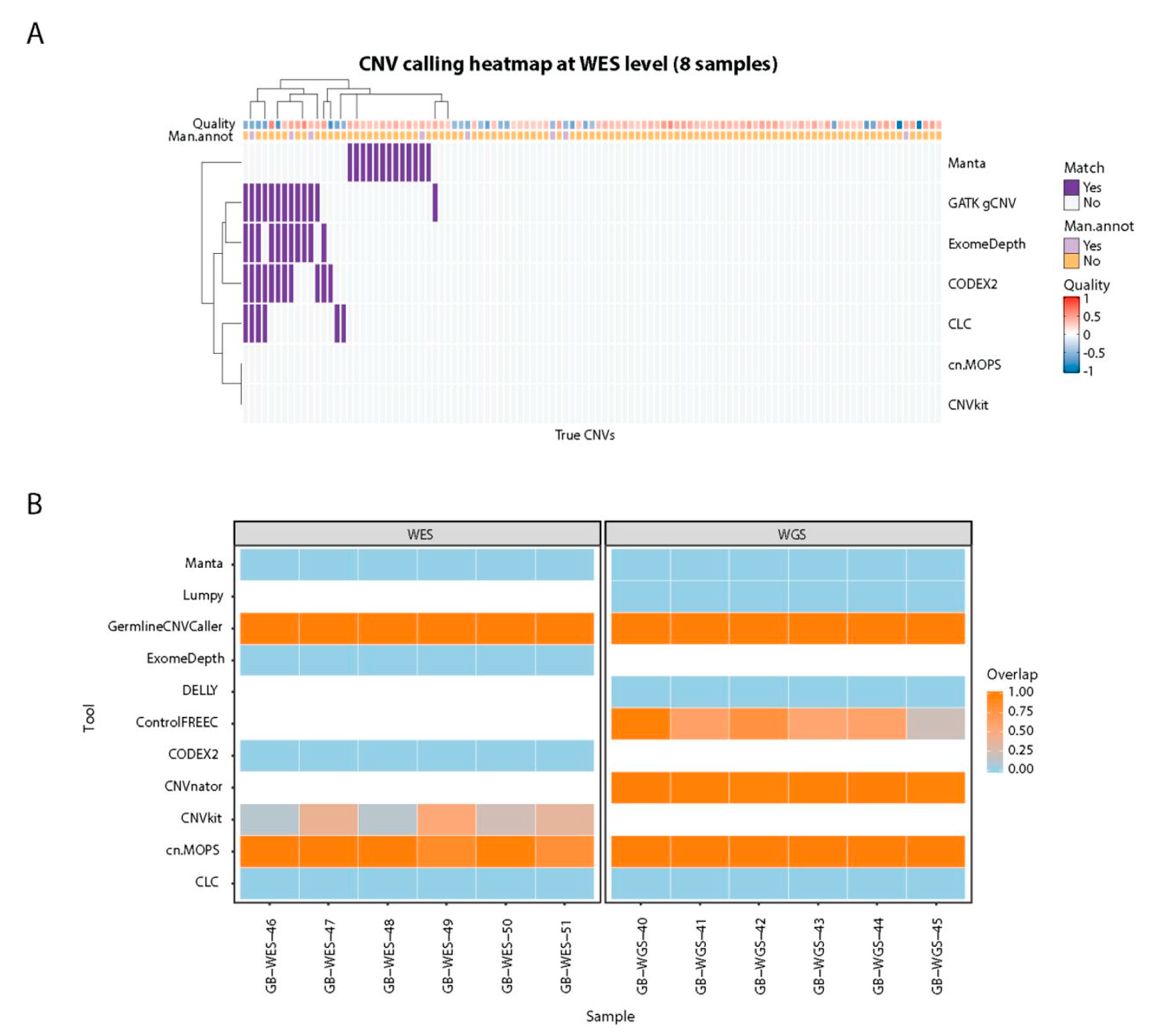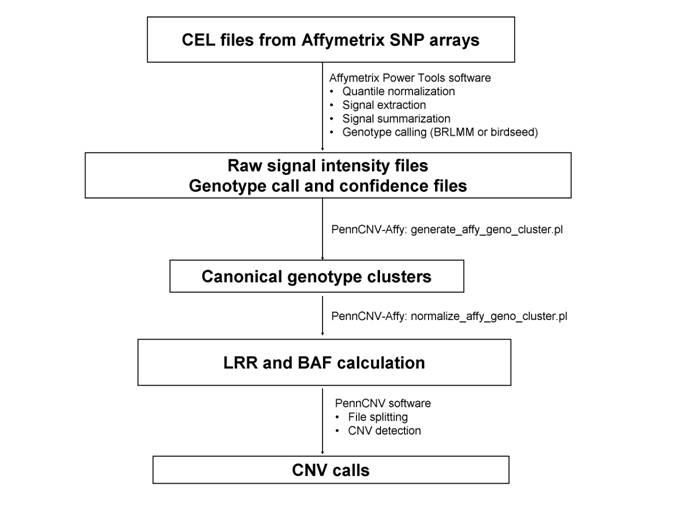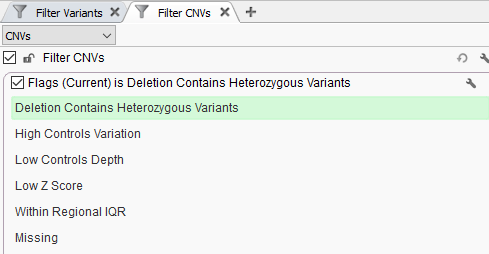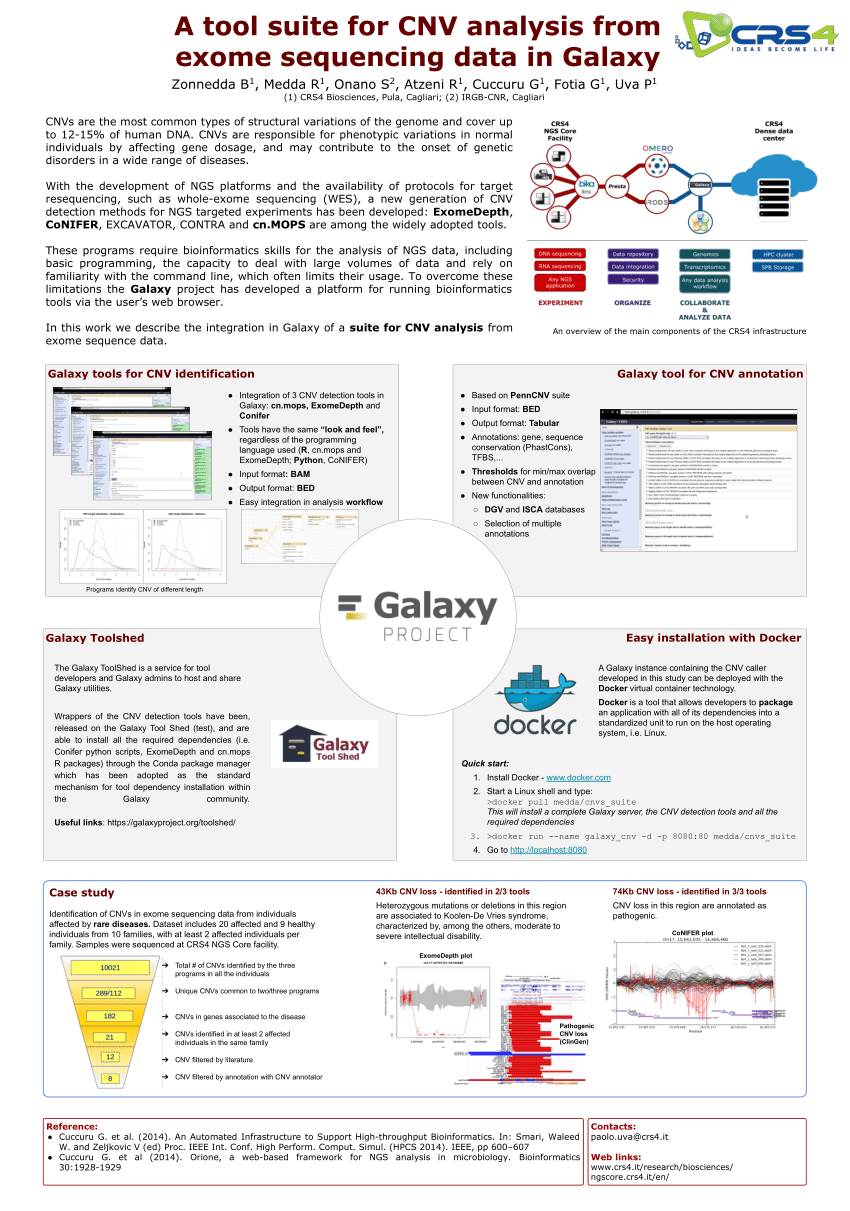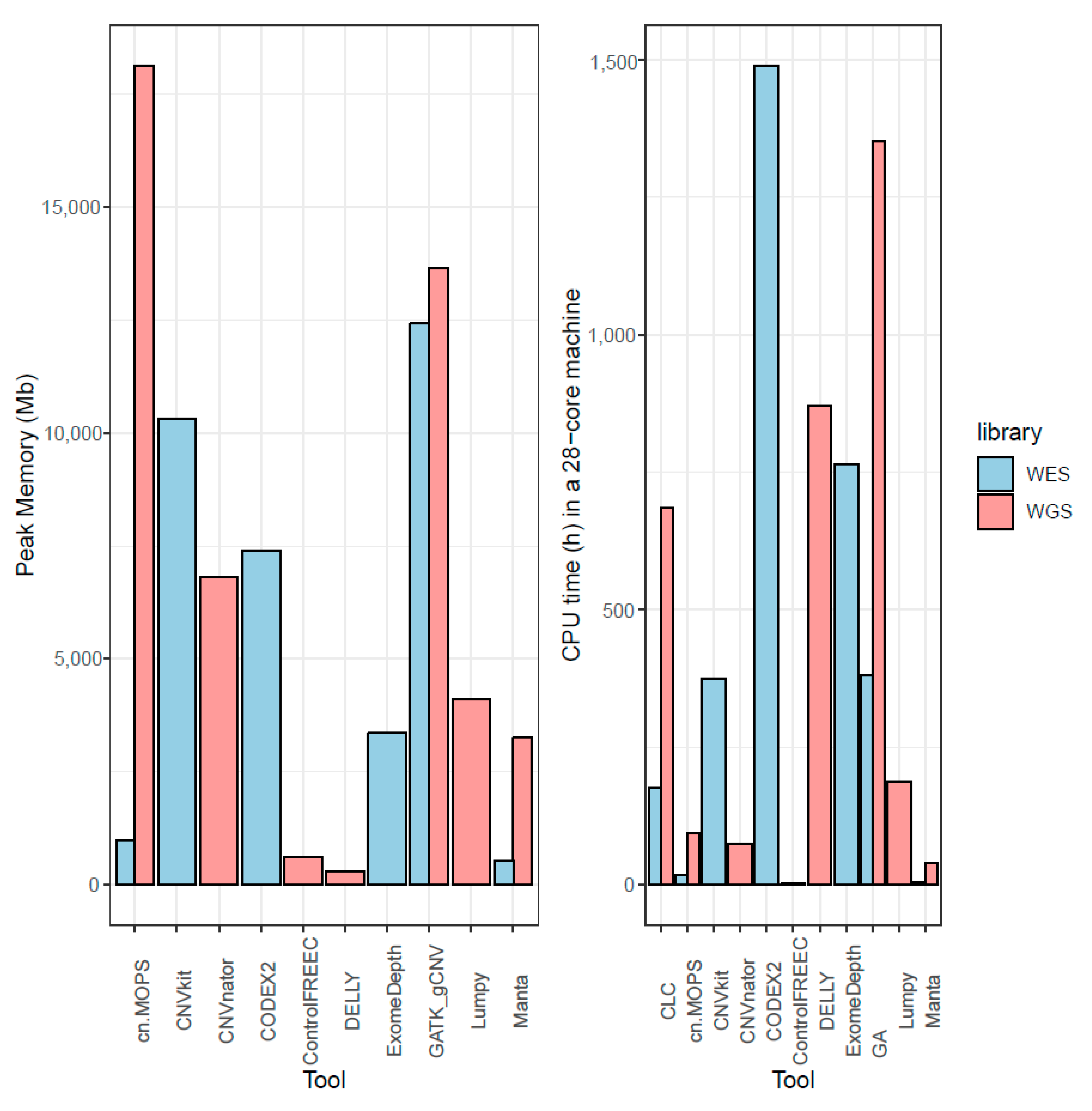
Cancers | Free Full-Text | A Comparison of Tools for Copy-Number Variation Detection in Germline Whole Exome and Whole Genome Sequencing Data
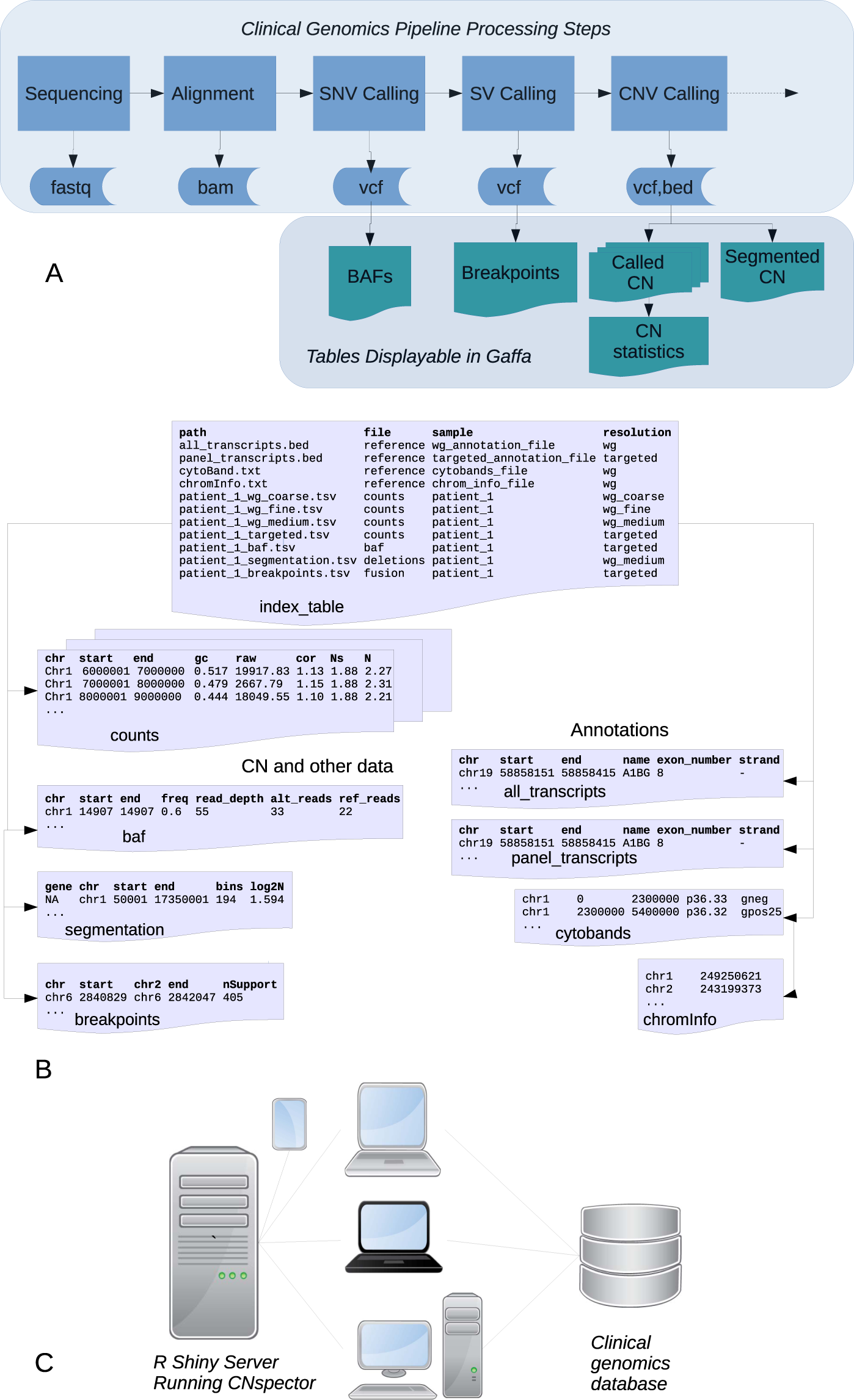
CNspector: a web-based tool for visualisation and clinical diagnosis of copy number variation from next generation sequencing | Scientific Reports

inCNV: An Integrated Analysis Tool for Copy Number Variation on Whole Exome Sequencing - Saowwapark Chanwigoon, Sakkayaphab Piwluang, Duangdao Wichadakul, 2020
New tools for CNV calling and low-frequency somatic calling · Issue #3322 · bcbio/bcbio-nextgen · GitHub

A) Number of duplications and deletions called by CNV calling tools in... | Download Scientific Diagram

Comparative study of whole exome sequencing-based copy number variation detection tools | BMC Bioinformatics | Full Text

The changes of tools' performances with respect to the CNV size. Fig a... | Download Scientific Diagram
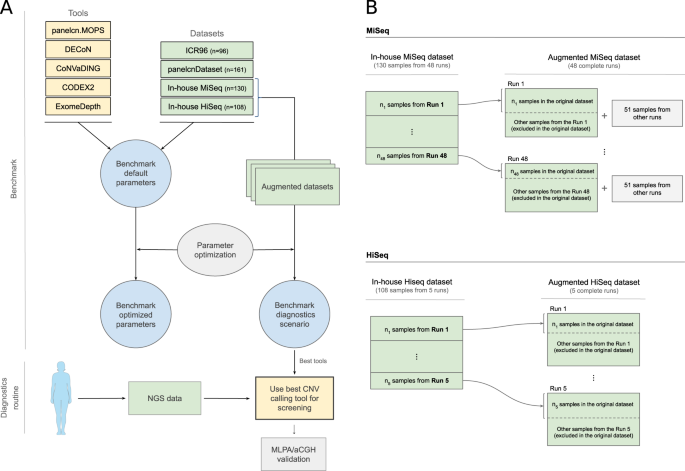
Evaluation of CNV detection tools for NGS panel data in genetic diagnostics | European Journal of Human Genetics
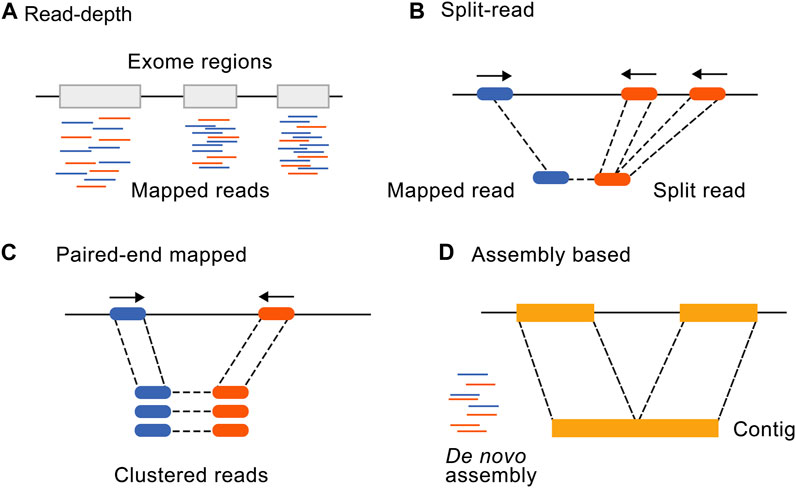
Frontiers | Incorporating CNV analysis improves the yield of exome sequencing for rare monogenic disorders—an important consideration for resource-constrained settings

Comparative study of whole exome sequencing-based copy number variation detection tools | BMC Bioinformatics | Full Text
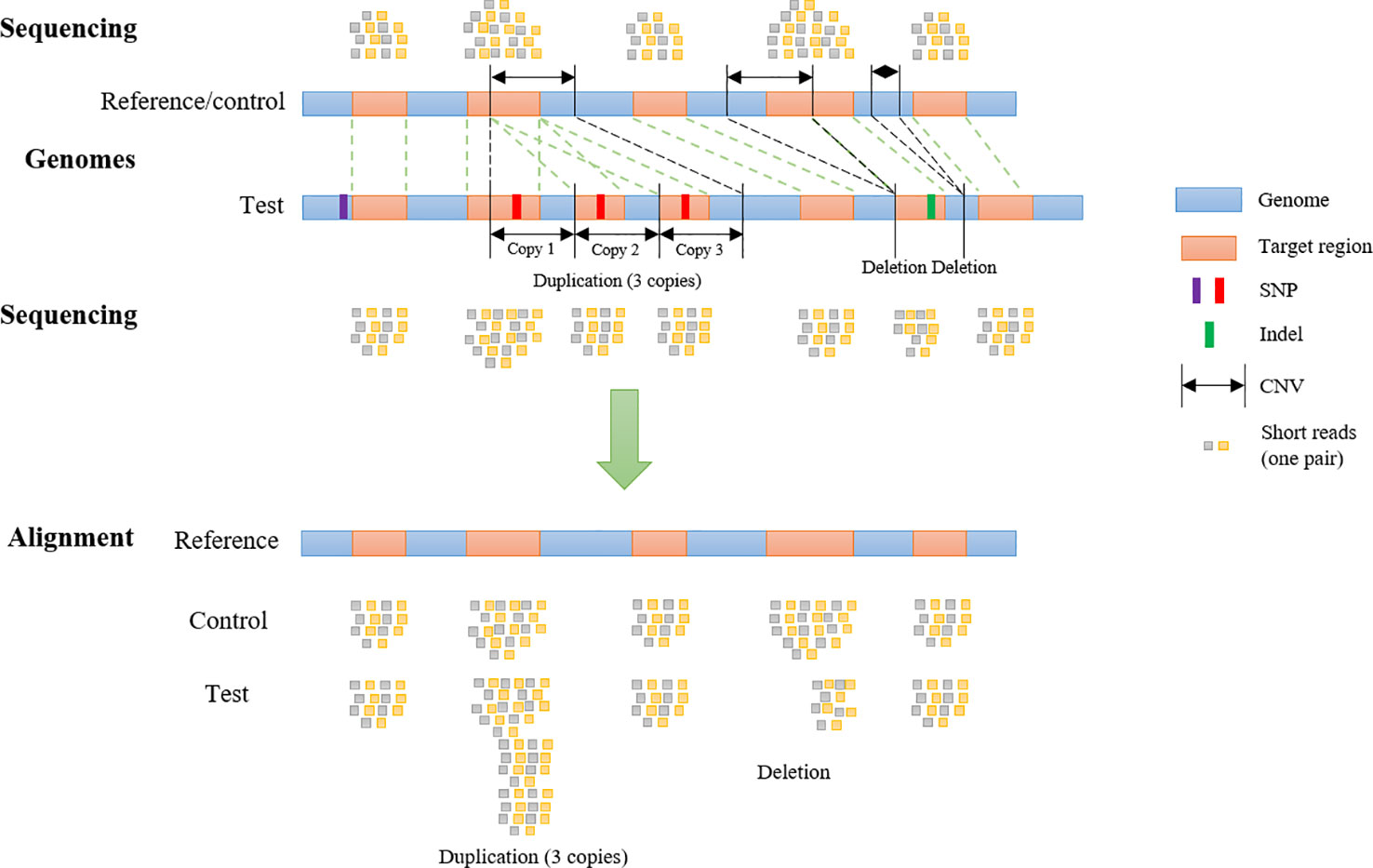
Frontiers | SECNVs: A Simulator of Copy Number Variants and Whole-Exome Sequences From Reference Genomes

inCNV: An Integrated Analysis Tool for Copy Number Variation on Whole Exome Sequencing - Saowwapark Chanwigoon, Sakkayaphab Piwluang, Duangdao Wichadakul, 2020
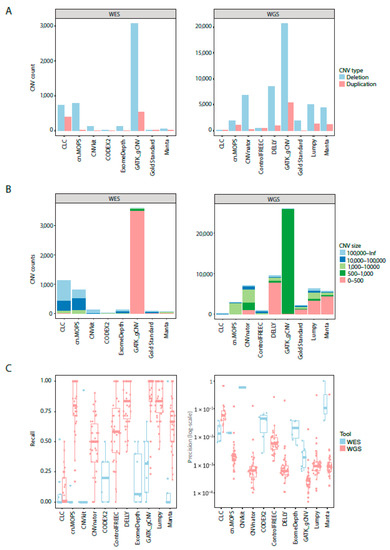
Cancers | Free Full-Text | A Comparison of Tools for Copy-Number Variation Detection in Germline Whole Exome and Whole Genome Sequencing Data